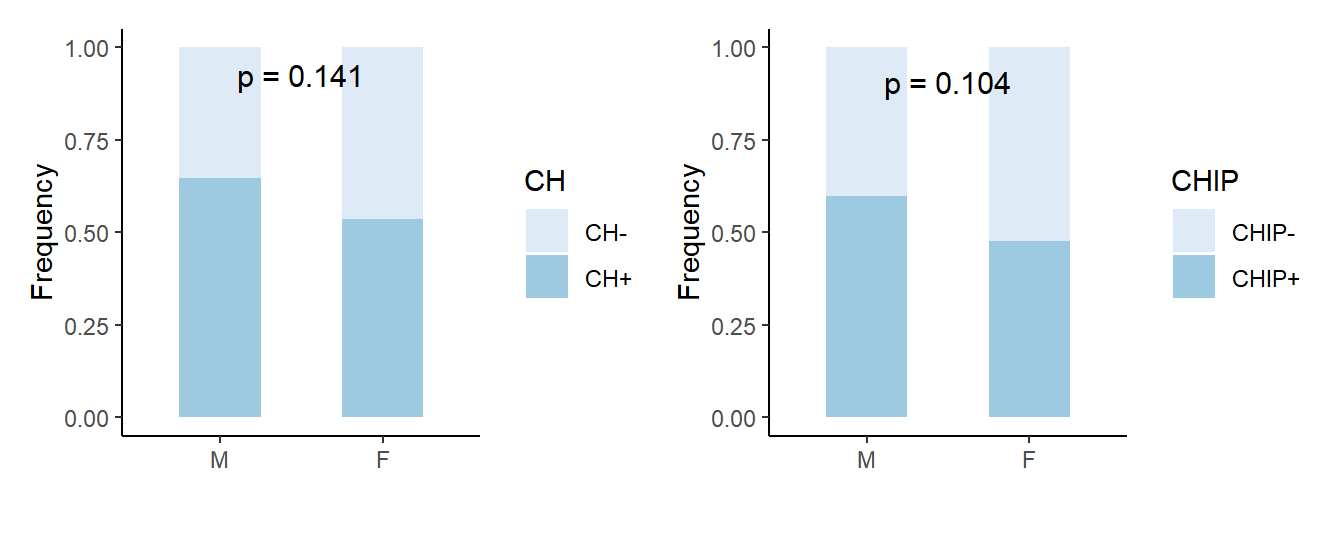
# prepare data (CH)
df_ch_all<- df_full %>% mutate(CH=ifelse(is.na(vaf),'N','Y')) %>% select(ID,Age,Sex,agrp,lngv,CH) %>% arrange(desc(CH)) %>% distinct(ID,Age,Sex,agrp,lngv,.keep_all=T) %>% mutate(Sex=factor(Sex,levels=c("M","F")))
df_ch1 <- df_ch_all%>% count(Sex,CH)
df_ch2 <- df_ch_all%>% count(Sex) %>% rename(total=n)
df_ch_num <-df_ch1 %>% inner_join(df_ch2,by="Sex") %>% mutate(freq=n/total) %>% mutate(CH=ifelse(CH=="Y","CH+","CH-"))
# plot Sex*CH
ch_sex <- df_ch_num %>% ggplot(aes(x=Sex,y=freq,fill=CH))+
geom_bar(stat="identity", width=0.5) + scale_fill_brewer(palette=1) + theme_classic() + xlab('') + ylab('Frequency')+
annotate("text", x = 1.5, y = max(df_ch_num$freq),
label = paste("p =", signif(chisq.test(table(df_ch_all$CH, df_ch_all$Sex))$p.value, digits = 3)),
vjust = -4, size = 4) +
theme_classic()
# prepare data (CHIP)
df_chip_all<- df_full %>% mutate(CHIP=ifelse(vaf<2,'N','Y')) %>% mutate(CHIP=ifelse(is.na(CHIP),"N",CHIP)) %>% select(ID,Age,Sex,agrp,lngv,CHIP,vaf) %>% arrange(desc(CHIP)) %>% distinct(ID,Age,Sex,agrp,lngv,.keep_all=T) %>% mutate(Sex=factor(Sex,levels=c("M","F")))
df_chip_1 <- df_chip_all%>% count(Sex,CHIP)
df_chip_2 <- df_chip_all%>% count(Sex) %>% rename(total=n)
df_chip_num <-df_chip_1 %>% inner_join(df_chip_2,by="Sex") %>% mutate(freq=n/total) %>% mutate(CHIP=ifelse(CHIP=="Y","CHIP+","CHIP-"))
# plot Sex*CHIP
chip_sex <- df_chip_num %>% ggplot(aes(x=Sex,y=freq,fill=CHIP)) +
geom_bar(stat="identity", width=0.5) + scale_fill_brewer(palette=1) + theme_classic() + xlab('') + ylab('Frequency')+
annotate(
"text", x = 1.5, y = max(df_chip_num$freq),
label = paste("p =", signif(chisq.test(table(df_chip_all$CHIP, df_chip_all$Sex))$p.value, digits = 3)),
vjust = -4.5, size = 4
) +
theme_classic()
# save into a file 'Supplemental_sex.pdf'
sex_both <- list(ch_sex,chip_sex) %>% wrap_plots(ncol=2, widths=c(1,1))
file.path(outdir,"Supplemental_sex.pdf") %>% ggsave(sex_both,width = 9,height=3)