6 Figure 5
Fig. 5: Independent validation of the clinical translational value of identified biomarkers in PDAC cohorts using IHC staining. (a-b) Representative IHC images of PDAC and TAC tissues stained for NDUFB8 (a) and CEMIP2 (b). (c-d) Kaplan-Meier survival curves comparing OS between patient subgroups stratified by adjuvant chemotherapy and the high/low expression of NDUFB8 (c) and CEMIP2 (d) in “RJ-cohort 3”. (e-f) Kaplan-Meier survival curves comparing OS between patient subgroups stratified by adjuvant chemotherapy and the high/low expression of NDUFB8 (e) and CEMIP2 (f) in “SH-cohort”. (g-h) Kaplan-Meier survival curves comparing OS between patient subgroups stratified by adjuvant chemotherapy and the high/low expression of NDUFB8 (g) and CEMIP2 (h) in “FR-cohort”. P-values were calculated using the log-rank test, and the hazard ratios calculated using the univariable Cox regression analysis.
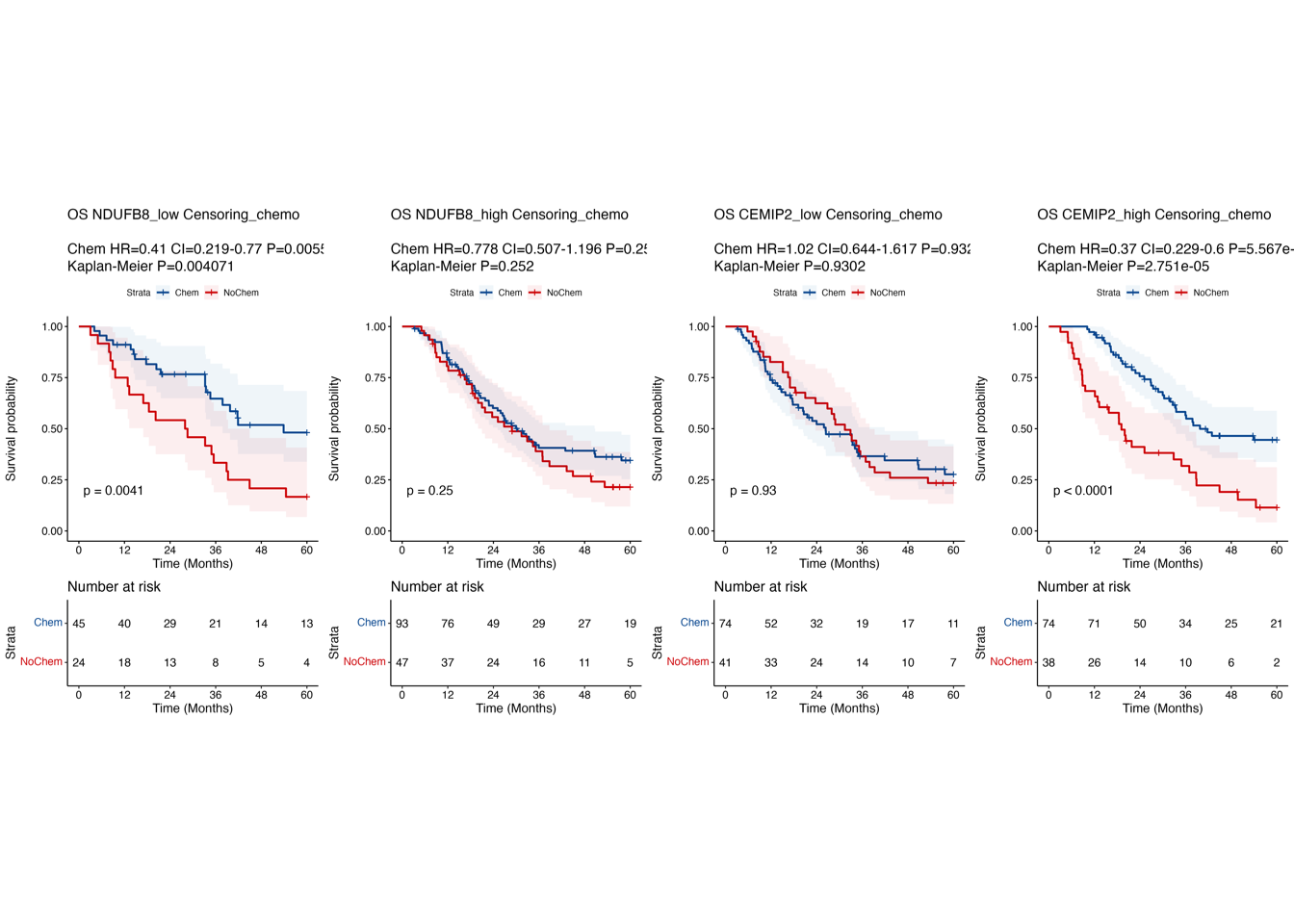
6.1 (cd) Kaplan-Meier curves in RJ-cohort 3
Kaplan-Meier survival curves comparing OS between patient subgroups stratified by adjuvant chemotherapy and the high/low expression of NDUFB8 (c) and CEMIP2 (d) in “RJ-cohort 3”.
dir.create("./results/Figure5")
#----------------------------------------------------------------------------------
# Step 1: Load the Data
#----------------------------------------------------------------------------------
# Sample information, clinical features and protein levels of markers in RJ-cohort 3
rj3.cohort <- read.xlsx("./data/Extended Data Table 7.xlsx", startRow = 2)
plot.cohort <- rj3.cohort %>%
mutate(Censoring_chemo_rev = Censoring_chemo,
Censoring_chemo = factor(as.character(Censoring_chemo), levels = c("Chem","NoChem")),
Censoring_chemo_rev = factor(as.character(Censoring_chemo_rev), levels = c("NoChem","Chem"))) %>%
mutate(NDUFB8_l = factor(as.character(NDUFB8_l), levels = c("Low","High")),
CEMIP2_l = factor(as.character(CEMIP2_l), levels = c("Low","High"))) %>%
mutate(GemType_rev = GemType,
GemType = factor(as.character(GemType), levels = c("Gem","NonGem","NoChem")),
GemType_rev = factor(as.character(GemType_rev), levels = c("NoChem","Gem","NonGem")))
summary(coxph(Surv(OS_month, OS_status) ~ Censoring_chemo_rev * NDUFB8_l, data = plot.cohort))
summary(coxph(Surv(OS_month, OS_status) ~ Censoring_chemo_rev * CEMIP2_l, data = plot.cohort))
idx <- which(plot.cohort$OS_month > 60)
plot.cohort$OS_month[idx] <- 60
plot.cohort$OS_status[idx] <- 0
#----------------------------------------------------------------------------------
# Step 2: OS survival stratified by lasso level in [NDUFB8 low] patients
#----------------------------------------------------------------------------------
plot.data <- plot.cohort[which(plot.cohort$NDUFB8_l == "Low"), ]
info <- summary(coxph(Surv(OS_month, OS_status) ~ Censoring_chemo_rev, data = plot.data))
anno.text <- ""
for (i in 1:nrow(info$conf.int)) {
anno.text <- paste0(anno.text, "\n", paste0(rownames(info$conf.int)[i], " HR=", round(info$conf.int[i, 1], 3), " CI=", round(info$conf.int[i, 3], 3), "-", round(info$conf.int[i, 4], 3), " P=", signif(info$coefficients[i, 5], 4) ))
}
anno.text <- paste0(anno.text, "\nKaplan-Meier P=", signif(survdiff(Surv(OS_month, OS_status) ~ Censoring_chemo_rev, data = plot.data)$pvalue, 4) )
anno.text <- str_replace_all(anno.text, "Censoring_chemo_rev", "")
fit <- survfit(Surv(OS_month, OS_status) ~ Censoring_chemo, data = plot.data)
p1 <- ggsurvplot(fit,
data = plot.data,
xlab = 'Time (Months)',
pval = TRUE,
risk.table = TRUE,
risk.table.height = 0.28,
legend.labs = c("Chem","NoChem"),
conf.int = TRUE, conf.int.alpha = 0.05,
palette = c("#00599F","#d80700"),
axes.offset = TRUE,
break.time.by = 12, xlim = c(0, 60),
title= paste0("OS NDUFB8_low Censoring_chemo \n", anno.text))
#----------------------------------------------------------------------------------
# Step 3: OS survival stratified by lasso level in [NDUFB8 High] patients
#----------------------------------------------------------------------------------
plot.data <- plot.cohort[which(plot.cohort$NDUFB8_l == "High"), ]
info <- summary(coxph(Surv(OS_month, OS_status) ~ Censoring_chemo_rev, data = plot.data))
anno.text <- ""
for (i in 1:nrow(info$conf.int)) {
anno.text <- paste0(anno.text, "\n", paste0(rownames(info$conf.int)[i], " HR=", round(info$conf.int[i, 1], 3), " CI=", round(info$conf.int[i, 3], 3), "-", round(info$conf.int[i, 4], 3), " P=", signif(info$coefficients[i, 5], 4) ))
}
anno.text <- paste0(anno.text, "\nKaplan-Meier P=", signif(survdiff(Surv(OS_month, OS_status) ~ Censoring_chemo_rev, data = plot.data)$pvalue, 4) )
anno.text <- str_replace_all(anno.text, "Censoring_chemo_rev", "")
fit <- survfit(Surv(OS_month, OS_status) ~ Censoring_chemo, data = plot.data)
p2 <- ggsurvplot(fit,
data = plot.data,
xlab = 'Time (Months)',
pval = TRUE,
risk.table = TRUE,
risk.table.height = 0.28,
legend.labs = c("Chem","NoChem"),
conf.int = TRUE, conf.int.alpha = 0.05,
palette = c("#00599F","#d80700"),
axes.offset = TRUE,
break.time.by = 12, xlim = c(0, 60),
title= paste0("OS NDUFB8_high Censoring_chemo \n", anno.text))
#----------------------------------------------------------------------------------
# Step 4: OS survival stratified by lasso level in [CEMIP2 low] patients
#----------------------------------------------------------------------------------
plot.data <- plot.cohort[which(plot.cohort$CEMIP2_l == "Low"), ]
info <- summary(coxph(Surv(OS_month, OS_status) ~ Censoring_chemo_rev, data = plot.data))
anno.text <- ""
for (i in 1:nrow(info$conf.int)) {
anno.text <- paste0(anno.text, "\n", paste0(rownames(info$conf.int)[i], " HR=", round(info$conf.int[i, 1], 3), " CI=", round(info$conf.int[i, 3], 3), "-", round(info$conf.int[i, 4], 3), " P=", signif(info$coefficients[i, 5], 4) ))
}
anno.text <- paste0(anno.text, "\nKaplan-Meier P=", signif(survdiff(Surv(OS_month, OS_status) ~ Censoring_chemo_rev, data = plot.data)$pvalue, 4) )
anno.text <- str_replace_all(anno.text, "Censoring_chemo_rev", "")
fit <- survfit(Surv(OS_month, OS_status) ~ Censoring_chemo, data = plot.data)
p3 <- ggsurvplot(fit,
data = plot.data,
xlab = 'Time (Months)',
pval = TRUE,
risk.table = TRUE,
risk.table.height = 0.28,
legend.labs = c("Chem","NoChem"),
conf.int = TRUE, conf.int.alpha = 0.05,
palette = c("#00599F","#d80700"),
axes.offset = TRUE,
break.time.by = 12, xlim = c(0, 60),
title= paste0("OS CEMIP2_low Censoring_chemo \n", anno.text))
#----------------------------------------------------------------------------------
# Step 5: OS survival stratified by lasso level in [CEMIP2 High] patients
#----------------------------------------------------------------------------------
plot.data <- plot.cohort[which(plot.cohort$CEMIP2_l == "High"), ]
info <- summary(coxph(Surv(OS_month, OS_status) ~ Censoring_chemo_rev, data = plot.data))
anno.text <- ""
for (i in 1:nrow(info$conf.int)) {
anno.text <- paste0(anno.text, "\n", paste0(rownames(info$conf.int)[i], " HR=", round(info$conf.int[i, 1], 3), " CI=", round(info$conf.int[i, 3], 3), "-", round(info$conf.int[i, 4], 3), " P=", signif(info$coefficients[i, 5], 4) ))
}
anno.text <- paste0(anno.text, "\nKaplan-Meier P=", signif(survdiff(Surv(OS_month, OS_status) ~ Censoring_chemo_rev, data = plot.data)$pvalue, 4) )
anno.text <- str_replace_all(anno.text, "Censoring_chemo_rev", "")
fit <- survfit(Surv(OS_month, OS_status) ~ Censoring_chemo, data = plot.data)
p4 <- ggsurvplot(fit,
data = plot.data,
xlab = 'Time (Months)',
pval = TRUE,
risk.table = TRUE,
risk.table.height = 0.28,
legend.labs = c("Chem","NoChem"),
conf.int = TRUE, conf.int.alpha = 0.05,
palette = c("#00599F","#d80700"),
axes.offset = TRUE,
break.time.by = 12, xlim = c(0, 60),
title= paste0("OS CEMIP2_high Censoring_chemo \n", anno.text))
# output
p <- arrange_ggsurvplots(list(p1, p2, p3, p4), ncol = 4, nrow = 1, print = FALSE)
ggsave(paste0("./results/Figure5/Figure5cd.pdf"), p, width = 20, height = 8)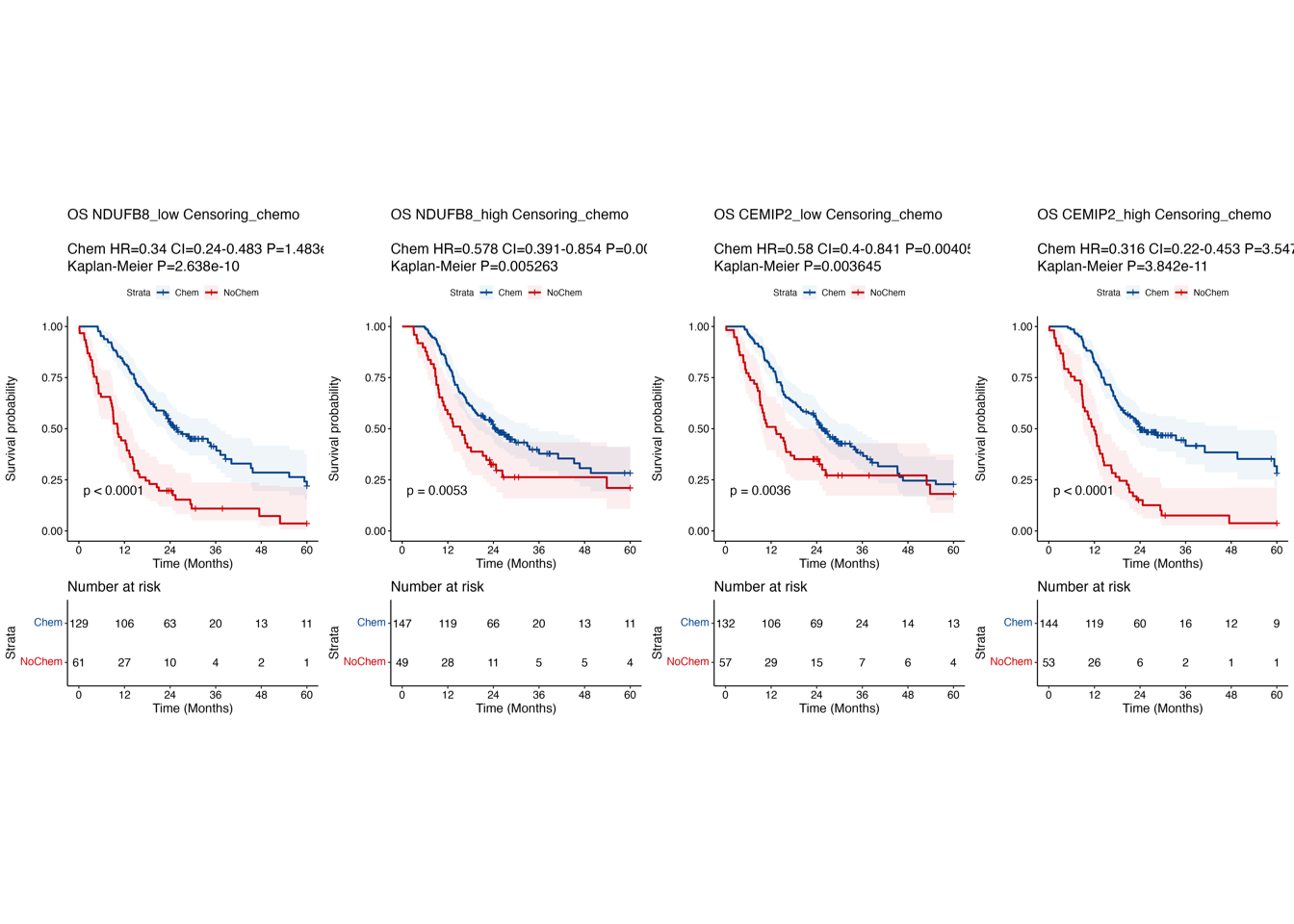
6.2 (ef) Kaplan-Meier curves in SH-cohort
Kaplan-Meier curves comparing OS between patient subgroups stratified by adjuvant chemotherapy and the high/low expression of NDUFB8 (e) and CEMIP2 (f) in “SH-cohort”.
#----------------------------------------------------------------------------------
# Step 1: Load the Data
#----------------------------------------------------------------------------------
# Sample information, clinical features and protein levels of markers in SH-cohort.
sh.cohort <- read.xlsx("./data/Extended Data Table 8.xlsx", startRow = 2)
plot.cohort <- sh.cohort %>%
mutate(Censoring_chemo_rev = Censoring_chemo,
Censoring_chemo = factor(as.character(Censoring_chemo), levels = c("Chem","NoChem")),
Censoring_chemo_rev = factor(as.character(Censoring_chemo_rev), levels = c("NoChem","Chem"))) %>%
mutate(NDUFB8_l = factor(as.character(NDUFB8_l), levels = c("Low","High")),
CEMIP2_l = factor(as.character(CEMIP2_l), levels = c("Low","High")))
summary(coxph(Surv(OS_month, OS_status) ~ Censoring_chemo_rev * NDUFB8_l, data = plot.cohort))
summary(coxph(Surv(OS_month, OS_status) ~ Censoring_chemo_rev * CEMIP2_l, data = plot.cohort))
idx <- which(plot.cohort$OS_month > 60)
plot.cohort$OS_month[idx] <- 60
plot.cohort$OS_status[idx] <- 0
#----------------------------------------------------------------------------------
# Step 2: OS survival stratified by lasso level in [NDUFB8 low] patients
#----------------------------------------------------------------------------------
plot.data <- plot.cohort[which(plot.cohort$NDUFB8_l == "Low"), ]
info <- summary(coxph(Surv(OS_month, OS_status) ~ Censoring_chemo_rev, data = plot.data))
anno.text <- ""
for (i in 1:nrow(info$conf.int)) {
anno.text <- paste0(anno.text, "\n", paste0(rownames(info$conf.int)[i], " HR=", round(info$conf.int[i, 1], 3), " CI=", round(info$conf.int[i, 3], 3), "-", round(info$conf.int[i, 4], 3), " P=", signif(info$coefficients[i, 5], 4) ))
}
anno.text <- paste0(anno.text, "\nKaplan-Meier P=", signif(survdiff(Surv(OS_month, OS_status) ~ Censoring_chemo_rev, data = plot.data)$pvalue, 4) )
anno.text <- str_replace_all(anno.text, "Censoring_chemo_rev", "")
fit <- survfit(Surv(OS_month, OS_status) ~ Censoring_chemo, data = plot.data)
p1 <- ggsurvplot(fit,
data = plot.data,
xlab = 'Time (Months)',
pval = TRUE,
risk.table = TRUE,
risk.table.height = 0.28,
legend.labs = c("Chem","NoChem"),
conf.int = TRUE, conf.int.alpha = 0.05,
palette = c("#00599F","#d80700"),
axes.offset = TRUE,
break.time.by = 12, xlim = c(0, 60),
title= paste0("OS NDUFB8_low Censoring_chemo \n", anno.text))
#----------------------------------------------------------------------------------
# Step 3: OS survival stratified by lasso level in [NDUFB8 High] patients
#----------------------------------------------------------------------------------
plot.data <- plot.cohort[which(plot.cohort$NDUFB8_l == "High"), ]
info <- summary(coxph(Surv(OS_month, OS_status) ~ Censoring_chemo_rev, data = plot.data))
anno.text <- ""
for (i in 1:nrow(info$conf.int)) {
anno.text <- paste0(anno.text, "\n", paste0(rownames(info$conf.int)[i], " HR=", round(info$conf.int[i, 1], 3), " CI=", round(info$conf.int[i, 3], 3), "-", round(info$conf.int[i, 4], 3), " P=", signif(info$coefficients[i, 5], 4) ))
}
anno.text <- paste0(anno.text, "\nKaplan-Meier P=", signif(survdiff(Surv(OS_month, OS_status) ~ Censoring_chemo_rev, data = plot.data)$pvalue, 4) )
anno.text <- str_replace_all(anno.text, "Censoring_chemo_rev", "")
fit <- survfit(Surv(OS_month, OS_status) ~ Censoring_chemo, data = plot.data)
p2 <- ggsurvplot(fit,
data = plot.data,
xlab = 'Time (Months)',
pval = TRUE,
risk.table = TRUE,
risk.table.height = 0.28,
legend.labs = c("Chem","NoChem"),
conf.int = TRUE, conf.int.alpha = 0.05,
palette = c("#00599F","#d80700"),
axes.offset = TRUE,
break.time.by = 12, xlim = c(0, 60),
title= paste0("OS NDUFB8_high Censoring_chemo \n", anno.text))
#----------------------------------------------------------------------------------
# Step 4: OS survival stratified by lasso level in [CEMIP2 low] patients
#----------------------------------------------------------------------------------
plot.data <- plot.cohort[which(plot.cohort$CEMIP2_l == "Low"), ]
info <- summary(coxph(Surv(OS_month, OS_status) ~ Censoring_chemo_rev, data = plot.data))
anno.text <- ""
for (i in 1:nrow(info$conf.int)) {
anno.text <- paste0(anno.text, "\n", paste0(rownames(info$conf.int)[i], " HR=", round(info$conf.int[i, 1], 3), " CI=", round(info$conf.int[i, 3], 3), "-", round(info$conf.int[i, 4], 3), " P=", signif(info$coefficients[i, 5], 4) ))
}
anno.text <- paste0(anno.text, "\nKaplan-Meier P=", signif(survdiff(Surv(OS_month, OS_status) ~ Censoring_chemo_rev, data = plot.data)$pvalue, 4) )
anno.text <- str_replace_all(anno.text, "Censoring_chemo_rev", "")
fit <- survfit(Surv(OS_month, OS_status) ~ Censoring_chemo, data = plot.data)
p3 <- ggsurvplot(fit,
data = plot.data,
xlab = 'Time (Months)',
pval = TRUE,
risk.table = TRUE,
risk.table.height = 0.28,
legend.labs = c("Chem","NoChem"),
conf.int = TRUE, conf.int.alpha = 0.05,
palette = c("#00599F","#d80700"),
axes.offset = TRUE,
break.time.by = 12, xlim = c(0, 60),
title= paste0("OS CEMIP2_low Censoring_chemo \n", anno.text))
#----------------------------------------------------------------------------------
# Step 5: OS survival stratified by lasso level in [CEMIP2 High] patients
#----------------------------------------------------------------------------------
plot.data <- plot.cohort[which(plot.cohort$CEMIP2_l == "High"), ]
info <- summary(coxph(Surv(OS_month, OS_status) ~ Censoring_chemo_rev, data = plot.data))
anno.text <- ""
for (i in 1:nrow(info$conf.int)) {
anno.text <- paste0(anno.text, "\n", paste0(rownames(info$conf.int)[i], " HR=", round(info$conf.int[i, 1], 3), " CI=", round(info$conf.int[i, 3], 3), "-", round(info$conf.int[i, 4], 3), " P=", signif(info$coefficients[i, 5], 4) ))
}
anno.text <- paste0(anno.text, "\nKaplan-Meier P=", signif(survdiff(Surv(OS_month, OS_status) ~ Censoring_chemo_rev, data = plot.data)$pvalue, 4) )
anno.text <- str_replace_all(anno.text, "Censoring_chemo_rev", "")
fit <- survfit(Surv(OS_month, OS_status) ~ Censoring_chemo, data = plot.data)
p4 <- ggsurvplot(fit,
data = plot.data,
xlab = 'Time (Months)',
pval = TRUE,
risk.table = TRUE,
risk.table.height = 0.28,
legend.labs = c("Chem","NoChem"),
conf.int = TRUE, conf.int.alpha = 0.05,
palette = c("#00599F","#d80700"),
axes.offset = TRUE,
break.time.by = 12, xlim = c(0, 60),
title= paste0("OS CEMIP2_high Censoring_chemo \n", anno.text))
# output
p <- arrange_ggsurvplots(list(p1, p2, p3, p4), ncol = 4, nrow = 1, print = FALSE)
ggsave(paste0("./results/Figure5/Figure5ef.pdf"), p, width = 20, height = 8)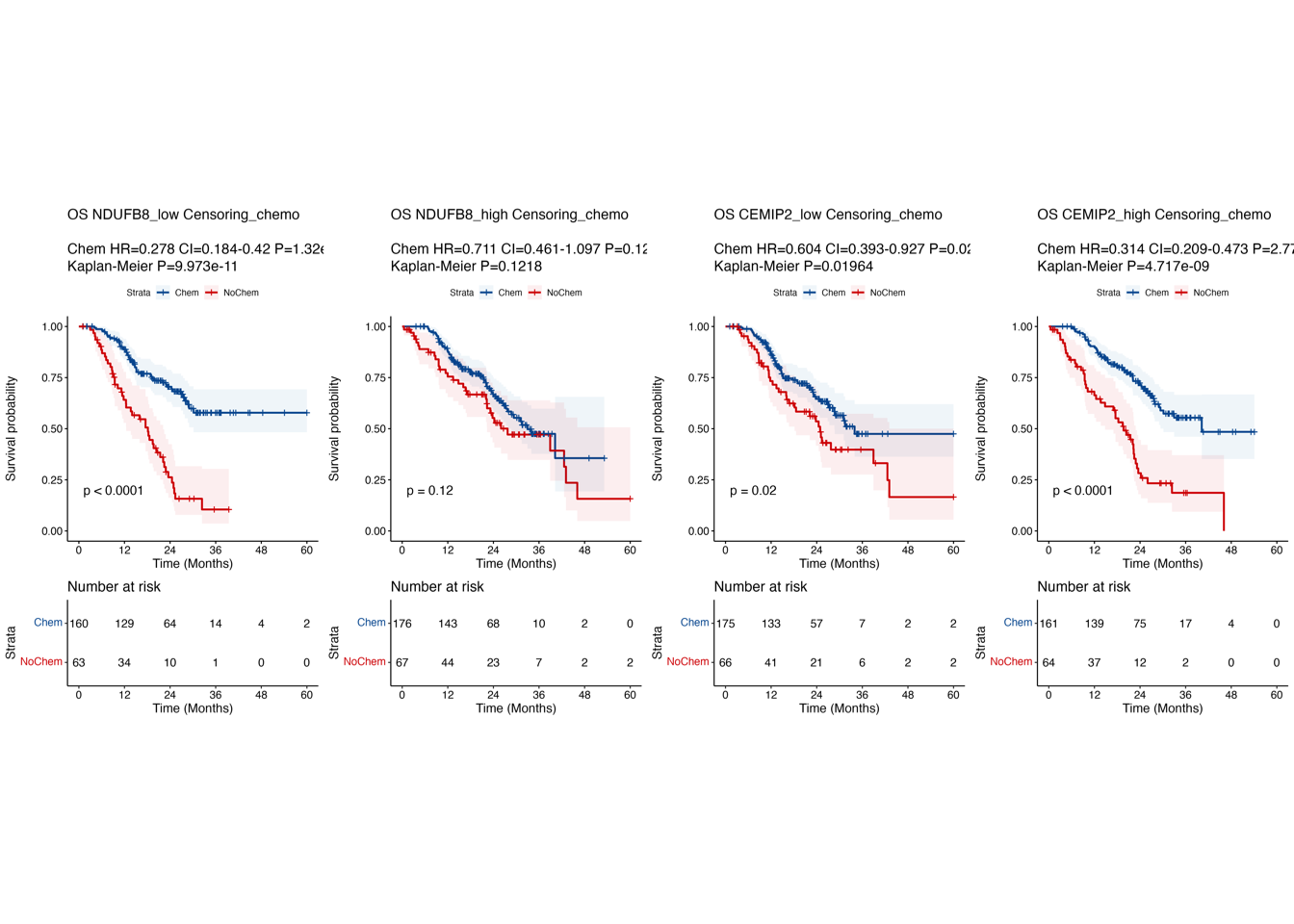
6.3 (gh) Kaplan-Meier curves in FR-cohort
Kaplan-Meier curves comparing OS between patient subgroups stratified by adjuvant chemotherapy and the high/low expression of NDUFB8 (g) and CEMIP2 (h) in “FR-cohort”. P-values were calculated using the log-rank test, and the hazard ratios calculated using the univariable Cox regression analysis.
#----------------------------------------------------------------------------------
# Step 1: Load the Data
#----------------------------------------------------------------------------------
# Sample information, clinical features and protein levels of markers in FR-cohort.
fr.cohort <- read.xlsx("./data/Extended Data Table 9.xlsx", startRow = 2)
plot.cohort <- fr.cohort %>%
mutate(Censoring_chemo_rev = Censoring_chemo,
Censoring_chemo = factor(as.character(Censoring_chemo), levels = c("Chem","NoChem")),
Censoring_chemo_rev = factor(as.character(Censoring_chemo_rev), levels = c("NoChem","Chem"))) %>%
mutate(NDUFB8_l = factor(as.character(NDUFB8_l), levels = c("Low","High")),
CEMIP2_l = factor(as.character(CEMIP2_l), levels = c("Low","High")))
summary(coxph(Surv(OS_month, OS_status) ~ Censoring_chemo_rev * NDUFB8_l, data = plot.cohort))
summary(coxph(Surv(OS_month, OS_status) ~ Censoring_chemo_rev * CEMIP2_l, data = plot.cohort))
idx <- which(plot.cohort$OS_month > 60)
plot.cohort$OS_month[idx] <- 60
plot.cohort$OS_status[idx] <- 0
#----------------------------------------------------------------------------------
# Step 2: OS survival stratified by lasso level in [NDUFB8 low] patients
#----------------------------------------------------------------------------------
plot.data <- plot.cohort[which(plot.cohort$NDUFB8_l == "Low"), ]
info <- summary(coxph(Surv(OS_month, OS_status) ~ Censoring_chemo_rev, data = plot.data))
anno.text <- ""
for (i in 1:nrow(info$conf.int)) {
anno.text <- paste0(anno.text, "\n", paste0(rownames(info$conf.int)[i], " HR=", round(info$conf.int[i, 1], 3), " CI=", round(info$conf.int[i, 3], 3), "-", round(info$conf.int[i, 4], 3), " P=", signif(info$coefficients[i, 5], 4) ))
}
anno.text <- paste0(anno.text, "\nKaplan-Meier P=", signif(survdiff(Surv(OS_month, OS_status) ~ Censoring_chemo_rev, data = plot.data)$pvalue, 4) )
anno.text <- str_replace_all(anno.text, "Censoring_chemo_rev", "")
fit <- survfit(Surv(OS_month, OS_status) ~ Censoring_chemo, data = plot.data)
p1 <- ggsurvplot(fit,
data = plot.data,
xlab = 'Time (Months)',
pval = TRUE,
risk.table = TRUE,
risk.table.height = 0.28,
legend.labs = c("Chem","NoChem"),
conf.int = TRUE, conf.int.alpha = 0.05,
palette = c("#00599F","#d80700"),
axes.offset = TRUE,
break.time.by = 12, xlim = c(0, 60),
title= paste0("OS NDUFB8_low Censoring_chemo \n", anno.text))
#----------------------------------------------------------------------------------
# Step 3: OS survival stratified by lasso level in [NDUFB8 High] patients
#----------------------------------------------------------------------------------
plot.data <- plot.cohort[which(plot.cohort$NDUFB8_l == "High"), ]
info <- summary(coxph(Surv(OS_month, OS_status) ~ Censoring_chemo_rev, data = plot.data))
anno.text <- ""
for (i in 1:nrow(info$conf.int)) {
anno.text <- paste0(anno.text, "\n", paste0(rownames(info$conf.int)[i], " HR=", round(info$conf.int[i, 1], 3), " CI=", round(info$conf.int[i, 3], 3), "-", round(info$conf.int[i, 4], 3), " P=", signif(info$coefficients[i, 5], 4) ))
}
anno.text <- paste0(anno.text, "\nKaplan-Meier P=", signif(survdiff(Surv(OS_month, OS_status) ~ Censoring_chemo_rev, data = plot.data)$pvalue, 4) )
anno.text <- str_replace_all(anno.text, "Censoring_chemo_rev", "")
fit <- survfit(Surv(OS_month, OS_status) ~ Censoring_chemo, data = plot.data)
p2 <- ggsurvplot(fit,
data = plot.data,
xlab = 'Time (Months)',
pval = TRUE,
risk.table = TRUE,
risk.table.height = 0.28,
legend.labs = c("Chem","NoChem"),
conf.int = TRUE, conf.int.alpha = 0.05,
palette = c("#00599F","#d80700"),
axes.offset = TRUE,
break.time.by = 12, xlim = c(0, 60),
title= paste0("OS NDUFB8_high Censoring_chemo \n", anno.text))
#----------------------------------------------------------------------------------
# Step 4: OS survival stratified by lasso level in [CEMIP2 low] patients
#----------------------------------------------------------------------------------
plot.data <- plot.cohort[which(plot.cohort$CEMIP2_l == "Low"), ]
info <- summary(coxph(Surv(OS_month, OS_status) ~ Censoring_chemo_rev, data = plot.data))
anno.text <- ""
for (i in 1:nrow(info$conf.int)) {
anno.text <- paste0(anno.text, "\n", paste0(rownames(info$conf.int)[i], " HR=", round(info$conf.int[i, 1], 3), " CI=", round(info$conf.int[i, 3], 3), "-", round(info$conf.int[i, 4], 3), " P=", signif(info$coefficients[i, 5], 4) ))
}
anno.text <- paste0(anno.text, "\nKaplan-Meier P=", signif(survdiff(Surv(OS_month, OS_status) ~ Censoring_chemo_rev, data = plot.data)$pvalue, 4) )
anno.text <- str_replace_all(anno.text, "Censoring_chemo_rev", "")
fit <- survfit(Surv(OS_month, OS_status) ~ Censoring_chemo, data = plot.data)
p3 <- ggsurvplot(fit,
data = plot.data,
xlab = 'Time (Months)',
pval = TRUE,
risk.table = TRUE,
risk.table.height = 0.28,
legend.labs = c("Chem","NoChem"),
conf.int = TRUE, conf.int.alpha = 0.05,
palette = c("#00599F","#d80700"),
axes.offset = TRUE,
break.time.by = 12, xlim = c(0, 60),
title= paste0("OS CEMIP2_low Censoring_chemo \n", anno.text))
#----------------------------------------------------------------------------------
# Step 5: OS survival stratified by lasso level in [CEMIP2 High] patients
#----------------------------------------------------------------------------------
plot.data <- plot.cohort[which(plot.cohort$CEMIP2_l == "High"), ]
info <- summary(coxph(Surv(OS_month, OS_status) ~ Censoring_chemo_rev, data = plot.data))
anno.text <- ""
for (i in 1:nrow(info$conf.int)) {
anno.text <- paste0(anno.text, "\n", paste0(rownames(info$conf.int)[i], " HR=", round(info$conf.int[i, 1], 3), " CI=", round(info$conf.int[i, 3], 3), "-", round(info$conf.int[i, 4], 3), " P=", signif(info$coefficients[i, 5], 4) ))
}
anno.text <- paste0(anno.text, "\nKaplan-Meier P=", signif(survdiff(Surv(OS_month, OS_status) ~ Censoring_chemo_rev, data = plot.data)$pvalue, 4) )
anno.text <- str_replace_all(anno.text, "Censoring_chemo_rev", "")
fit <- survfit(Surv(OS_month, OS_status) ~ Censoring_chemo, data = plot.data)
p4 <- ggsurvplot(fit,
data = plot.data,
xlab = 'Time (Months)',
pval = TRUE,
risk.table = TRUE,
risk.table.height = 0.28,
legend.labs = c("Chem","NoChem"),
conf.int = TRUE, conf.int.alpha = 0.05,
palette = c("#00599F","#d80700"),
axes.offset = TRUE,
break.time.by = 12, xlim = c(0, 60),
title= paste0("OS CEMIP2_high Censoring_chemo \n", anno.text))
# output
p <- arrange_ggsurvplots(list(p1, p2, p3, p4), ncol = 4, nrow = 1, print = FALSE)
ggsave(paste0("./results/Figure5/Figure5gh.pdf"), p, width = 20, height = 8)